Published in PLOS ONE, 2021.
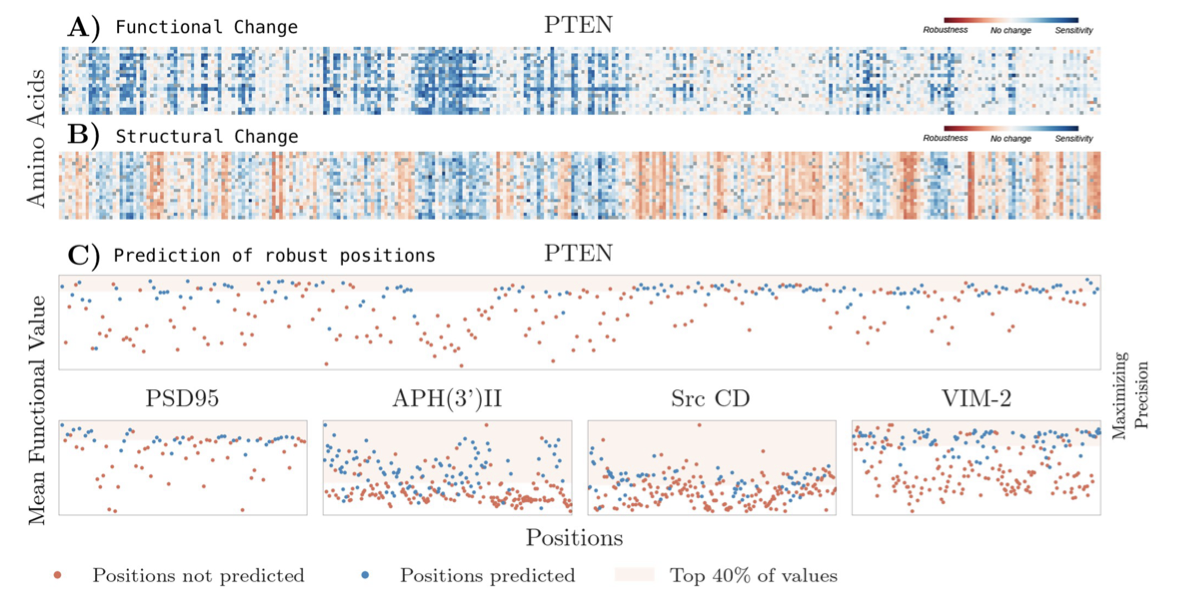
This project looks into the structural and functional shifts in proteins caused by mutations, exploring how such changes impact protein behavior. By constructing detailed amino acid networks for five proteins, we analyzed how mutations at specific positions alter both structure and function, revealing a strong relationship between structural sensitivity and functional outcomes. Our approach combines network science with protein modeling, allowing us to predict functionally sensitive regions based solely on structural perturbations.
We developed a computational framework that uses network-based measures, such as the size and connectivity of perturbation networks, to assess the structural impacts of mutations. The results suggest that structural robustness often aligns with functional stability, while pronounced structural shifts tend to disrupt protein function. Our model achieved notable precision and recall, offering a valuable tool for predicting mutation-sensitive positions in proteins and shedding light on how structure-driven mutations may guide protein evolution and stability.